...the collection, classification, storage, and analysis of biochemical and biological information using computers especially as applied to molecular genetics and genomics (Merriam Webster).
 |
| Apologies to Disney and MicrobeWiki |
Sometimes we want to know which tiny species of bacteria are inhabiting a particular ecosystem. Maybe we want to know what bacteria make us healthy, or maybe we want to know what bacteria just killed a dozen diners. Maybe we just want to see who is living in our bellybutton. Join me as I take you on a journey in "Finding Bifido."
There are several ways to discover microbial life. The easiest is by looking at a droplet containing microbes under a powerful microscope and comparing what you see to a picture, much like picking out a mugger in a lineup down at the 12th Precinct.
 |
| NatGeo |
 |
| Microscopic image of gram-negative Pseudomonas aeruginosa bacteria (pink-red rods) |
By examining how a bacteria reacts to dye as well as how it behaves, grows, and dies, researchers have done a great job finding bacteria for the last couple hundred years. Finding bacteria in this manner requires that the bacteria be "culturable," unfortunately, only a small percentage of bacteria are willing to be grown in a Petri dish. A scanning electron micrograph imager lets the researcher see an individual bacteria:
 |
| Wikipedia |
DNA
Bacteria are living creatures. They have DNA, just like all lifeforms, and this DNA can give clues to exactly what species of bacteria we hold in our hand.
Up until the '90's, DNA was a simple concept. It was shown as a double-helix made up of bonding pairs of chemicals labeled ACT and G. We now know that DNA can take many shapes and be made of one, three, and even four strands.
 |
| Scientific American |
We can solve crimes using the clues left behind by a tiny drop of DNA!
 |
| Berkeley.edu |
 |
| Daily Mail |
16s rRNA
DNA can be used to compare a couple of samples, but it generally contains simply too much information to be useful. RNA is a product of DNA. Sections of DNA ("genes"), produce copies of themselves called RNA. These RNA strands are single chains, not double-helix, but they are still made of the same chemicals we earlier labeled A, C, T, and G.
Note: At this point we could veer off into never-never land and discuss RNA until the end of time. Science is still learning about RNA and all its forms...don't ever let anyone fool you into believing that we know everything about DNA and RNA. The "junk DNA" of the '80's is the "non-coding Piwi-interacting micro RNA" of 2015.
Ribosomal RNA (rRNA) is a special molecule found in the ribosomes of the cells of all living things. rRNA is an ancient structure, and as such, preserves the identity of its owner very well. A section of rRNA known affectionately as "16s" is a highly conserved region found in the rRNA of all lifeforms and is commonly used to quickly identify bacteria.
16s rRNA can be identified in each bacteria by it's strings of ACTG. Every 16s rRNA chain contains approximately 1500 letters. Laid out flat, it looks like this:
 |
| PNAS |
And in 3D:
 |
| Wikipedia |
If this is starting to seem immensely complex, it's not. To a computer geek and a microbiologist, this presents no problem at all!
Identifying Bacteria
The first step in finding 16s rRNA involves chopping the bacteria up into tiny little pieces to extract its DNA and RNA. This chopping is done using standard chemicals found in biotech supply houses, and even on Amazon!
Chemicals used in this process:
- TE buffer: It is a buffer used for the storage of nucleic acids (DNA and RNA), and also to prevent it from degradation.
- Lysozyme: Enzyme that is used for degrading the cell wall of the organism.
- SDS: SDS is strong anionic detergent that helps in solubilizing the proteins and lipids present in the membranes. It exposes the chromosomes that contain the DNA, also helps in releasing DNA from histones and other DNA binding proteins by denaturing them.
- Proteinase K: Degrades most of the protein impurities in the DNA(Deproteination).
- Phenol: Helps in removing most of the protein impurities from the DNA.
- Chloroform: Prevents the shearing effect of DNA during the extraction process.
- Isoamyl alcohol: Reduce the formation of the foams in extraction techniques.
- Sodium acetate: Precipitates DNA in the solution
- Absolute ethanol: Precipitate the DNA out of solution.
The next step in identifying bacteria from this mash-up of "gut bug guts" is known as Polymerase Chain Reaction (PCR) amplification. PCR amplification is simple, but would take too long to describe. It was invented in 1983, and garnered a Nobel Prize for its inventor in 1993. Here again, a couple of chemicals and a few pieces of lab equipment, and viola!
Every piece of DNA chopped up in the extraction process, and subjected to 30 cycles of PCR, produces about one billion copies of each strand. The "primers" used ensure only segments of 16s rRNA are selected. Easy peasy!
At the end of the PCR process, we have a vial filled with segments of 16s rRNA that are random chains roughly 150 "letters" long. The fluid containing all these random segments is sorted using various techniques involving electrical current and certain gels that will quickly stratify the chains of 16s rRNA into a format that can then be read by a computer. The final step looks like this:
 |
| OpenI |
Bioinformatics
Biology meets information meets mathematics.
The field of bioinformatics has exploded in the past 20 years. What scientists spent years doing, aided by university supercomputers, can now be done at home in seconds on the crudest of home computers.
16s rRNA extraction culminates in the creation of a computer file. This file has a standard designation ".fasta". These fasta files are used widely in bioinformatics, but for the purposes of bacterial identification using 16s rRNA, they all look similar. If you open a fasta file, you'll see a ">" symbol followed by a whole bunch of A's, C's, T's and G's. These fasta files can be modifed to contain quality information (known as .fastq) or amino acid sequences (".faa").
The field of bioinformatics has exploded in the past 20 years. What scientists spent years doing, aided by university supercomputers, can now be done at home in seconds on the crudest of home computers.
16s rRNA extraction culminates in the creation of a computer file. This file has a standard designation ".fasta". These fasta files are used widely in bioinformatics, but for the purposes of bacterial identification using 16s rRNA, they all look similar. If you open a fasta file, you'll see a ">" symbol followed by a whole bunch of A's, C's, T's and G's. These fasta files can be modifed to contain quality information (known as .fastq) or amino acid sequences (".faa").
Finding Bifido
Companies that perform bacterial analysis using 16s rRNA sequencing such as uBiome or American Gut Project, provide a detailed analysis of samples of skin, mouth, genital, or fecal samples that patrons provide (for a fee!).
These companies will give a detailed report of what the sample contained, some companies also provide the fasta (or fastq) files associated with your report, called "raw data.". These reports can be a source of endless fun for the bioninformatician in you!
Once downloaded and unzipped, you'll find a standard fasta file. To open this file, you'll need to go to a free service like Galaxy.
When converted to a format you'll recognize, you'll see thousands and thousands of lines of ACTG's.
The start of each sequence is denoted by a ">", like this:
Let's Find Bifido!
Once downloaded and unzipped, you'll find a standard fasta file. To open this file, you'll need to go to a free service like Galaxy.
When converted to a format you'll recognize, you'll see thousands and thousands of lines of ACTG's.
The start of each sequence is denoted by a ">", like this:
>NS500457:47:H25CFAFXX:2:11101:10270:1118 1:N:0:GAGTGTGCCAGCAGCCGCGGTAATACGTAGGGTGCAATCGTTATCCGGAATTATTGGGCGTAAAGGGCTCGTAGGCGGTTCGTCGCGTCCGGTGTGAAAGTCCATCGCTTAACGGTGGCTCCGCGCCGGGTACGGGCGGGCTTGCGGGCGGT
The first stretch of numbers and letters (>NS500457:47:H25CFAFXX:2:11101:10270:1118 1:N:0:) is information about this sample and how it was processed. Computer programs understand this...you don't have to.
There are also free programs that will match this sequence against repositories of sequences that researchers all over the world have so kindly donated for the sake of science. Once such free program is Genome. Just copy the fasta sequence, choose a few options that denote RNA and which database you'd like to use, and click "Compute." In just a few seconds, you be provided a list of species that this sequence closely resembles. The sequence above results in 500 BLAST "hits". The #1 most likely suspect, Bifido!:
There are also free programs that will match this sequence against repositories of sequences that researchers all over the world have so kindly donated for the sake of science. Once such free program is Genome. Just copy the fasta sequence, choose a few options that denote RNA and which database you'd like to use, and click "Compute." In just a few seconds, you be provided a list of species that this sequence closely resembles. The sequence above results in 500 BLAST "hits". The #1 most likely suspect, Bifido!:
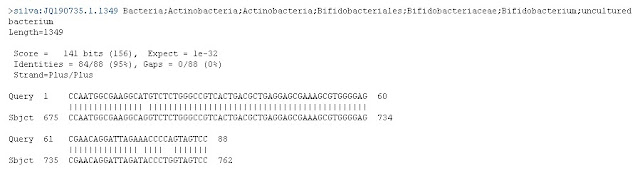
This graphic shows that our sequence of approximately 150 letters (Query) matched almost perfectly from position 1 through 94 to a "Subject." The subject's code is at least 618 letters long, and the match was in the region from 525 through 618. By clicking on the file highlighted in the BLAST results page, we see that this is the full sequence we believe we have a match with:
>JQ190735.1.1349 Bacteria;Actinobacteria;Actinobacteria;Bifidobacteriales; GAUGAACGCUGGCGGCGUGCUUAACACAUGCAAGUCGAACGGGAUCGGCUGGAGCUUGCUCCGGCCGUGAGAGUGGCGAA CGGGUGAGUAAUGCGUGACCGACCUGCCCCAUACACCGGAAUAGCUCCUGGAAACGGGUGGUAAUGCCGGAUGCUCCAGU UGGAUGCAUGUCCUUCUGGGAAAGAUUUAUCGGUAUGGGAUGGGGUCGCGUCCUAUCAGGUAGUCGGCGGGGUAACGGCC CACCGAGCCUACGACGGGUAGCCGGCCUGAGAGGGCGACCGGCCACAUUGGGACUGAGAUACGGCCCAGACUCCUACGGG AGGCAGCAGUGGGGAAUAUUGCACAAUGGGCGCAAGCCUGAUGCAGCGACGCCGCGUGCGGGAUGACGGCCUUCGGGUUG UAAACCGCUUUUGACUGGGAGCAAGCCCUUCGGGGUGAGUGUACCUUUCGAAUAAGCACCGGCUAACUACGUGCCAGCAG CCGCGGUAAUACGUAGGGUGCAAGCGUUAUCCGGAAUUAUUGGGCGUAAAGGGCUCGUAGGCGGUUCGUCGCGUCCGGUG UGAAAGUCCAUCGCUUAACGGUGGAUCCGCGCCGGGUACGGGCGGGCUUGAGUGCGGUAGGGGAGACUGGAAUUCCCGGU GUAACGGUGGAAUGUGUAGAUAUCGGGAAGAACACCAAUGGCGAAGGCAGGUCUCUGGGCCGUCACUGACGCUGAGGAGC GAAAGCGUGGGGAGCGAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGGUGGAUGCUGGAUGUGGGGACCAUUC CACGGUCUCCGUGUCGGAGCCAACGCGUUAAGCAUCCCGCCUGGGGAGUACGGCCGCAAGGCUAAAACUCAAAGAAAUUG ACGGGGGCCCGCACAAGCGGCGGAGCAUGCGGAUUAAUUCGAUGCAACGCGAAGAACCUUACCUGGGCUUGACAUGUUCC CGACAGCCCCAGAGAUGGGGCCUCCCUUCGGGGCGGGUUCACAGGUGGUGCAUGGUCGUCGUCAGCUCGUGUCGUGAGAU GUUGGGUUAAGUCCCGCAACGAGCGCAACCCUCGCCCUGUGUUGCCAGCACGUCAUGGUGGGAACUCACGGGGGACCGCC GGGGUCAACUCGGAGGAAGGUGGGGAUGACGUCAGAUCAUCAUGCCCCUUACGUCCAGGGCUUCACGCAUGCUACAAUGG CCGGUACAACGGGAUGCGACCUCGUGAGGGGGAGCGGAUCCCUUAAAACCGGUCUCAGUUCGGAUUGGAGUCUGCAACCC GACUCCAUGAAGGCGGAGUCGCUAGUAAUCGCGGAUCAGCAACGCCGCGGUGAAUGCGUUCCCGGGCCU
Notice anything funny?
That's right! The "T's" have all been replaced with "U's" in this file. Some researchers use a U instead of a T when discussing RNA. BLAST recognizes this and didn't miss a beat.
Another strange thing you may notice is that the #1 BLAST hit is to an "uncultured" Bifido species. If we want to drill in even deeper, we need more information!
Reverse fasta
RNA sequences always proceed in an orderly fashion, however, as they are produced in a cyclone of whirling chemical reactions (Fasta! Fasta!), no one can say if the end result of 150 letters is presented forward, backward, or even in mirror-image. Some of the providers of these reports also give the raw data in a reverse fasta file. This reverse file can be identified by having the exact same "informational text" in the very first line, but differentiated slightly in some way to show it's a reverse of the first. This reverse file is not a simple reversing of the letter sequence, but a completely different analysis of the first sample, done in reverse, so it may result in a completely different outcome when BLASTed. Easier to just watch!
The reverse fasta of our "Finding Bifido" sequence looks like this (Obtained by uploading raw data fasta file into Galaxy):
>NS500457:47:H25CFAFXX:2:11101:10270:1118 2:N:0:GACACCGGACTACTGGGGTTTCTAATCCTGTTCGCTCCCCACGCTTTCGCTCCTCAGGTCAGTGACGGCCCAGAGACATGCCTTCGCCATTGGAGTACTTCCCGATATCTACACATTCCACCGTTACACCGGGAATTCCAGTCTCCCCTA
The only difference on the informational line is the "2" (highlighted in pink). The sequence that follows the descriptor is a totally new set of ACTG, which will we want to BLAST, resulting in this #1 hit:
This is the exact same match that the first sequence got us, but do you notice anything different? The first sequence that we BLASTed, resulted in a sequence match from letters 1 through 94 of our "query" using letters 525 through 618 of the subject. In this sequence match, the matching letters were found in letters 618 through 711, and they are listed backwards! This is a big clue that our second file is, indeed, a reverse fasta.
Reverse-complementing
"My, you sure look ugly today!" Ooops, not that kind of reverse compliment?
In reverse-complementing, we need to look at the sequence in mirror image, and also switch the ACTGs to match their natural binding partners. Sounds complicated, but it's not. There's an app for that!
Back in Galaxy, we find an option to "reverse-complement:" our sequence, the reverse complement of the above file looks like this:
>NS500457:47:H25CFAFXX:2:11101:10270:1118 2:N:0:TAGGGGAGACTGGAATTCCCGGTGTAACGGTGGAATGTGTAGATATCGGGAAGTACTCCAATGGCGAAGGCATGTCTCTGGGCCGTCACTGACGCTGAGGAGCGAAAGCGTGGGGAGCGAACAGGATTAGAAACCCCAGTAGTCCGGTGTC
In this version, everything in the informational line is the same (note the pink "2"), but all of the C's are now G's and the A's are now T's and the new sequence is backwards. Confirm this by looking at the first 5 letters of the original sequence: " GACAC" and comparing them with their mirror image in the last 5 letters of the new sequence: "GTGTC." Get it? Simple! Mirror imagery: GTGTC|GACAC
Don't worry, some people's brains don't see things like this...that's why there's an app. Can you imagine doing that by hand?
So what if we BLAST this reverse-complemented file? We get this:
It's the same exact file we've been getting, still at #1, but now with a bit less confidence. The first 2 BLASTs were shown as 97% indentity, this one is 95%. Also some of the sequence numbers are a bit shorter. However, we now see in the "Subject" sequence, that the order has again been reversed, meaning that by "reverse-complementing" this file, we have lined it up with the original sequence.
Joining Files
Here comes the fun part! Until now, we've been dealing with sequences 150 letters long, and only using 80 or 90 of the letters in the #1 BLAST hits. What if we simply join these two files? Again, there are apps for that, but we can also easily do this manually by cutting and pasting one to the other, resulting in a 300 letter sequence:
GACACCGGACTACTGGGGTTTCTAATCCTGTTCGCTCCCCACGCTTTCGCTCCTCAGCGTCAGTGACGGCCCAGAGACATGCCTTCGCCATTGGAGTACTTCCCGATATCTACACATTCCACCGTTACACCGGGAATTCCAGTCTCCCCTATAGGGGAGACTGGAATTCCCGGTGTAACGGTGGAATGTGTAGATATCGGGAAGTACTCCAATGGCGAAGGCATGTCTCTGGGCCGTCACTGACGCTGAGGAGCGAAAGCGTGGGGAGCGAACAGGATTAGAAACCCCAGTAGTCCGGTGTC
And BLASTed:
Finally, we end up with a different result. This last BLAST, formed by reverse complementing the reverse file and joining it with the forward file, resulted in a 96% match with a different file, and this sequence shows that we are looking at the species Bifidobacteria adolescentis. I believe we just "Found Bifido!"
Were the extra steps worth it to discover which species of Bifido we were looking at? I believe so. Some sequencial analyses are not so straight forward, result in top 5 hits of completely different families or phyla of bacteria. It just so happens that Bifidobacteria is very well studied and lots of researchers have entered their databases into public repositories for all to use.
In some regards, knowing the bacterial species may not be as important as knowing the genus or even phylum. In case you've forgotten, a small refresher in taxonomy:
In some regards, knowing the bacterial species may not be as important as knowing the genus or even phylum. In case you've forgotten, a small refresher in taxonomy:
 |
| Citylab |
Conclusion
I hope you enjoyed this journey to Finding Bifido. The advances in science, biology, computers, and deciphering all this information is simply mind-boggling. As computers get faster and databases of bacteria grow larger, researchers will find new ways of determining exactly what bacteria call your body "home." For now, the science is still a bit young and requires a certain amount of guess-work, even when using computers.
For the advanced biohacker, the steps are like this (Thanks, Barney!):
For the advanced biohacker, the steps are like this (Thanks, Barney!):
Option 1 :
1. Combine all four R1 Lanes into a single file.
2. Combine all four R2 Lanes into a single file (Same lane order as step 1).
3. Reverse compliment the combined R2 file using a system like Galaxy or
equivalent. If using Fastq format, handle the quality appropriately.
4. Join the sequences from Step 1 with the reverse complimented
sequences from Step 3, sequence by sequence, to create a final file.
Each sequence will have a length equal to the length of the R1 sequence
+ the reverse complimented R2 sequence. Use the header from the R1
sequence for the joined sequence.
Note : To allow a system like MG-rast to handle quality filtering, use
Fastq format.
Option 2 :
1. Reverse compliment each of the four R2 lane files using a system like
Galaxy or equivalent. If using Fastq format, handle the quality
appropriately.
2. For each lane, join the sequences from the R1 file with the reverse
complimented R2 file from Step 1, sequence by sequence, to create a
final file. Each sequence will have a length equal to the length of the
R1 sequence + the reverse complimented R2 sequence. Use the header from
the R1 sequence for the joined sequence.
3. Combine the four lane files from Step 2 into a single file.
Note : To allow a system like MG-rast to handle quality filtering, use
Fastq format.
Questions?
Later!
Tim



Going back to the last time I took any Bio -- sophomore year of college. But I thought "U" coded for uracil, which is distinct from thyme and took its place in RNA?
ReplyDeleteSuppose it's been some years and my science may be out of date?
Thymine...mobile phones and autocorrect are sapping our collective brains.
ReplyDeleteWow! You read the post. I'm impressed. See, Barney! Told ya somebody would read it.
ReplyDeleteThymine and Uracil is a real mess. Yes, generally when discussing DNA, Thymine is the standard base and in RNA Uracil takes its place. However, when discussing RNA before it has encoded proteins, Thymine is still considered as the standard base.
Some places call it 16s DNA. Further, when using PCR amplification, RNA can "flow backwards" and get turned into complimentary DNA (cDNA).
You made my day. Thanks for asking! I actually just asked Dr. Ayers this exact same question last week! I was concerned that when a BLAST hit turned up a sequence that used U instead of T it would cause a low percentage, but it does not seem to be the case.
I'm a constant reader if not commenter =) I loved the information theory of genetics and gene expression in high school and college. Still do, as reading through here confirms!
DeleteI started reading the article on "junk DNA," and all the linked articles...Gonna lose some time to wiki-lag on this stuff!
I'm pleased I asked a relevant question, and fascinated to see this stuff is wonderfully more complicated than we learned in school...even college level survey courses. I actually considered biostatistics and bioinformatics as graduate programs when I was finishing my bachelor's, but ultimately decided against more school altogether.
Thanks for posting, always a pleasure to read!
I'm happy to have lost this bet! It was fun working with the raw data files. "Citizen science" can go beyond donating a sample and waiting for the results. With the raw data, you can dig down a little deeper, play with the free analytics, and learn more along the way. I hope you will enjoy it Padraic.
DeleteBarney
Padraic, et al. - Some interesting things to Google: "non-coding RNA," "G-Quadruplex," and "DNA argonaute". None of these things were known when I first took biology in the '90s. Or at least not talked about.
DeleteThis blog post is not a method for analyzing a uBiome report, it would take years doing it this way. Barney and I were dissecting fasta files and I had all these notes/sequences left from the hundreds of emails that little exercise generated, so I thought it would be fun to preserve them here, in a blog post.
The last couple paragraphs are Barney's notes on how to analyze a full uBiome sample on a batch-scale.
Nice post, thanks Tim and Barney!
ReplyDeleteBut I miss a tribute to Carl Woese, the scientist who realized "that DNA could unlock the hidden relationships between different organisms" and had the lucky idea to choose16s rRNA to identify prokaryotic species. Reading about him you also realize the power of IT, because back in the late 1960s he did all the work mentioned above manually...He also discovered a new domain of life - Archaea, and surprise, as it happens to novel theories and findings - nobody believed him, he grew bitter, felt rejected, and it took decades for his work to be recognized.
Some excerpts from:
The Man Who Rewrote the Tree of Life (2014)
"Carl Woese may be the greatest scientist you’ve never heard of. “Woese is to biology what Einstein is to physics,” says Norman Pace, a microbiologist at the University of Colorado, Boulder. A physicist-turned-microbiologist, Woese specialized in the fundamental molecules of life—nucleic acids—but his ambitions were hardly microscopic. He wanted to create a family tree of all life on Earth.
To create his evolutionary tree of life, then, Woese would need to choose a gene that was present in every known organism, one that was copied from generation to generation with a high degree of precision and mutated very slowly, so he would be able to track it over billions of years of evolution.
Some of the most ancient genes are those coding for molecules known as ribosomal RNAs. In ribosomes, parts of the cell that float around the soupy cytoplasm, proteins and ribosomal RNA, or rRNA, work together to crank out proteins. Each ribosome is composed of large and small subunits, which are similar in both simple, single-celled prokaryotes and more complex eukaryotes. Woese had several different rRNA molecules to choose from in the various subunits, which are classified based on their length. At around 120 nucleotides long, 5S rRNA wasn’t big enough to use to compare lots of different organisms. On the other end of the spectrum, 23S rRNA was more than 2300 nucleotides long, making it far too difficult for Woese to sequence using the technologies of the time. The Goldilocks molecule—long enough to allow for meaningful comparisons but not too long and difficult to sequence—was 16S rRNA in prokaryotes and its slightly longer eukaryotic equivalent, 18S rRNA. Woese decided to use these to create his quantitative tree of life."
(...)
Now, when scientists try to discover unknown microbial species, the first gene they sequence is 16S rRNA. “It’s become one of the fundamentals of biology,” Wolfe says. “After more than 20 years, Woese was finally vindicated.”
Woese died on December 30, 2012, at the age of 84 of complications from pancreatic cancer. At the time of his death, he had won some of biology’s most prestigious awards and had become one of the field’s most respected scientists. Thanks to Woese’s legacy, we now know that most of the world’s biodiversity is hidden from view, among the tiny microbes that live unseen in and around us, and in them, the story of how life first evolved on this planet."
Nice. Thank you for the perspective and context. IT seems a natural fit and helps to handle the large amounts of data.
DeleteBarney
This is very interesting! I wonder if it was a mistake using 16s versus a longer stretch now that we have computers to deal with the information?
DeleteA sequence 150 or 300 bits long is such a laughably small amount of data.
I am thoroughly impressed and confused. I appreciate all the information presented here and all that everyone posts to try to help educate us get us healthier, but most of this is way above my pay grade!
ReplyDeleteThanks,
Gina
+1
Delete+ me too! But will try going thru it with fresh brain in morning for what crumbs I can glean
Delete++++1
DeleteTim, please, learn to write, nobody understands you...junk DNA, OMG.
You should learn from Carl Zimmer and explain things sort of... better.
Is Most of Our DNA Garbage? (2015)
"T. Ryan Gregory’s lab at the University of Guelph in Ontario is a sort of genomic menagerie, stocked with creatures, living and dead, waiting to have their DNA laid bare.
Gregory’s investigations into all these genomes has taught him a big lesson about life: At its most fundamental level, it’s a mess
His favorite way to demonstrate this is through what he calls the “onion test,” which involves comparing the size of an onion’s genome to that of a human.
(...)
It showed that the onion’s genome was five times bigger than mine.
“The onion wins,” Gregory said. The onion always does.
But why? Why does an onion carry around so much more genetic material than a human? "
Happy, Wildcucumber?
Delighted, Gemma!
Delete"Gregory’s investigations into all these genomes has taught him a big lesson about life: At its most fundamental level, it’s a mess"
ha! I love that quote.
Most things in the scientific world that seem to be set in stone are generally, really, a mess.
Delete"The human genome contains around 20,000 genes, that is, the stretches of DNA that encode proteins. But these genes account for only about 1.2 percent of the total genome. The other 98.8 percent is known as noncoding DNA."
DeleteLet's add in the genome of all the bacteria that reside in, and on, us...the onion won't stand a chance!
As Gemma will remind you, the onion contains all these bacteria as well. Plus fungi and probably also viruses. The onion stands.
DeleteTim, you can pre-order: http://www.amazon.com/Thing-Explainer-Complicated-Stuff-Simple/dp/0544668251
DeleteGreat book about the matter "Junk DNA. A Journey Through the Dark Matter of the Genome." by Nessa Carey.
ReplyDeleteIf I remember right, in the talk by Rob Knight: How our microbes make us who we are. TED talk, he mentions about whole microbe genome sequencing. This will be done when it is affordable enough. Also, he said in Donna Gate's summit (http://healthygutsummit.com/) that every human has almost every gut microbe but these can't be detected with 16S but with whole genome sequencing. So that way with prebiotics you could change your microbiome without probiotics. Well, that is obvious to the readers of this blog.
In addition, when I did gut microbiome sequencing, the sequence provider had just found out that the color coded tag sequences that they were using to separate different samples in one sequencing reaction was masking some microbes and that way giving wrong results. So, we really need the whole microbiome sequencing to become less expensive and then we have problem of too much data or not.
Noora
Hi guys, I apoliogise since my question isn't related to the topic of a post, but I am in a "bacteria" conundrum and since Google didn't help much, I couldn't think of a better place to ask. I am dealing with IBS-D/SIBO-D, and have already been on strong herbal antimicrobials for couple of months... I just had a flare up and that means another 2 months minimum. Most practitioners are against probiotics while trying to get rid of SIBO overgrowth, but I can't fathom continuing with this "killing" regimen without supporting microform, I know I need it. Can you pls advise me probiotic products, or at least bacteria strain, that are safe for this situation and don't turn into D-Lactate in the body? this was the recommendation I got from a few sources.
ReplyDeleteWith gut dysbiosis firmly in place, it would be foolhardy for any of us to make recommendations. There are simply too many variables.
DeleteWhat caused the latest flare-up, any idea? "Strong herbal antimicrobials" isn't necessarily a "killing regimen" for the bacteria that will eventually become your healthy microbiome. I'm glad you did not say that you were on round after round of antibiotics.
I hesitate to give any advice, not because I am afraid of litigation, but because I have no idea what is causing your problems. It could be completely unrelated to the bacteria.
If I could help you based on the few lines you provided, I'd be a millionaire (or at least a very popular guy!).
I would recommend working with real doctors (not internet gurus) and giving them a couple months to fix you. If they can't or you don't like their advice, find a new one. In the end, you need to stop the constant annoyance and health problems caused by IBS-D, which may or may not be from SIBO.
After exhausting everything, you may want to see a "normal" doc who will prescribe PPIs. I have seen lots of people clear up years-long "SIBO" with a day or two of Nexium, indicating to me that SIBO was never the problem.
If PPIs clear you up rapidly, wean off them slowly and see what "breaks" you again. Most people that start PPIs cannot get off of them, however, or don't want to. But there are problems that doctors just cannot fix easily and these problems often get lumped into a finding of "SIBO" whether real or not.
If you find a doctor, naturopath or conventional, that can 100% say with testing that you have SIBO, it would go a long way to creating a treatment path. If they are just saying "SIBO" without knowing for sure, then this could be a long path to recovery.
At some point in this, probiotics such as Elixa, AOR3, PrescriptAssist, and others might help out.
Hope that helps!
Maybe others have better advice.
Tim
Kira - Could you tell me what these 'strong herbal antimicrobials' are and in what form you're taking them? I work with plant medicines and the current trend toward using herbs in this way interests me.
DeleteDear Tim, thank you for your detailed write up, you really gave me a few good thinking points... Regarding probiotics that don't turn into D-Lacate, do you have any idea where should I look for the info?
DeleteWildcucumber, the standard herbal antimicrobial treatment for SIBO is a choice between Allimed (Allicin), Berberine, Neem, Oregano Oil, Peppermint oil...the protocol should combine some of them to be taken for a month then you reassess, and if needed - take another month of a different combination from this list. There are products in pro lines of supplements that already provided certain combination, and lots of practitioners rely on these. Right now I am taking this product i found through research, its called MycoZyme by Apex Energetics (broad spectrum), as well as Neem and Allimed. Its amazing to me that just 2 days of MycoZyme stops 4 days worth of horrible the D. and painful gut cramps - in just 1-2 days! It must mean something as far as diagnosis goes that I react so quickly to it... But I digress... Pls LMK if you have any other questions;-)
DeleteKira - Thanks, it's just an academic interest so that was plenty to go on. Good luck on your journey.
Delete"Regarding probiotics that don't turn into D-Lacate, do you have any idea where should I look for the info? "
DeleteI have never given this any thought, bit I did find some papers that may have an answer for you:
http://www.pulsus.com/journals/abstract.jsp?sCurrPg=journal&jnlKy=2&atlKy=3854&isuKy=250&isArt=t [click on the "full text pdf" to see full]
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3723183/
" After the failure of antibiotic treatment, a stand-alone synbiotic treatment was started, specifically Bifidobacterium breve Yakult and Lactobacillus casei Shirota as probiotics, and galacto-oligosaccharide as a prebiotic. Serum D-lactate levels declined, and the patient has been recurrence-free for 3 years without dietary restriction. Synbiotics allowed the reduction in colonic absorption of D-lactate by both prevention of D-lactate–producing bacterial overgrowth and stimulation of intestinal motility, leading to remission of D-lactate acidosis."
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3999946/
"Intake of L. reuteri-containing formula was safe and did not cause an increase in d-lactic acid beyond two weeks."
So, possibly for you, a probiotic containing Lactobacillus reuteri and/or Bifido breve would be OK. Maybe others, if you read the papers, follow the references and similar papers to learn more.
Not a recommendation, but this is advertised as "D-lactate Free"
Deletehttp://www.customprobiotics.com/custom-probiotics-d-lactate.htm
thnx Tim! I also found one more that fits the bill: http://www.corganic.com/gutpro
DeleteHey Tim,some good news about good ol' bananas. apparently bananas should be eaten with the skin as they are packed with extra fiber and various bioactive compounds like polyphenols, carotenoids and others..
ReplyDeletehttp://www.dailymail.co.uk/health/article-3281298/Is-banana-SKIN-new-superfood-Peel-packed-nutrients-vitamins-aid-weight-loss-boost-mood.html
'The peel is also high in the mood-boosting hormone serotonin - a neurotransmitter derived from tryptophan.'
Deletenice!
Chimps don't peel bananas.
DeleteNot surprising when you see the teeth on a chimp.
Deletehttp://www.arkive.org/chimpanzee/pan-troglodytes/image-G4529.html
Banana peels are rather astringent. Banana flowers more so.
DeleteCurious about this. My internet research (fwiw) indicates some do, some don't, even among the same species. Also, I learned some time ago that chimps peel bananas differently from most humans - starting from the pointy end, not the stem. I'd guess that the type of banana matters too. The typical grocery store banana is easy to peel, but I've gotten some exotics that are nearly impossible even after deemed ripe. If I were hungry enough, I'd give in and just eat the whole thing rather than fighting. I'm not opposed to eating the peel, but it's not something that greatly ap-peels to me. Ha, ha.
DeleteOkay, so I'm posting the results of my most recent uBiome results. These were done September 2015.
ReplyDelete1
Bacteroides 32.19%
2
Blautia 15.83%
3
Roseburia 10.91%
4
Faecalibacterium 6.85%
5
Dorea 3.64%
6
Pseudobutyrivibrio 3.59%
7
Subdoligranulum 2.99%
8
Enterococcus 2.46%
9
Kluyvera 2.2%
10
Anaerotruncus 1.72%
11
Anaerostipes 1.49%
12
Clostridium 1.49%
13
Collinsella 1.33%
14
Sutterella 1.03%
15
Parabacteroides 0.89%
16
Paraprevotella 0.75%
17
Streptococcus 0.73%
18
Alistipes 0.66%
19
Barnesiella 0.66%
20
Sarcina 0.66%
21
Bifidobacterium 0.6%
22
Bilophila 0.47%
23
Lachnospira 0.42%
24
Phascolarctobacterium 0.28%
25
Flavonifractor 0.23%
26
Erysipelatoclostridium 0.2%
27
Hespellia 0.18%
28
Butyricimonas 0.13%
29
Intestinimonas 0.08%
30
Parasutterella 0.07%
31
Intestinibacter 0.04%
32
Enterorhabdus 0.04%
33
Haemophilus 0.04%
34
Enterobacter 0.03%
35
Holdemania 0.03%
36
Slackia 0.02%
37
Pantoea 0.02%
38
Granulicatella 0.01%
39
Weissella 0.01%
40
Candidatus Soleaferrea 0.01%
41
Acetanaerobacterium 0.01%
42
Gordonibacter 0.01%
43
Pseudoflavonifractor 0.01%
44
Peptostreptococcus 0.01%
45
Catabacter 0.01%
46
Megasphaera 0.01%
47
Prevotella 0.0%
48
Turicibacter 0.0%
49
Anaerofustis 0.0%
50
Anaerofilum 0.0%
51
Veillonella 0.0%
52
Peptoniphilus 0.0%
53
Dielma 0.0%
54
Marvinbryantia 0.0%
55
Terrisporobacter 0.0%
56
Pediococcus 0.0%
57
Moryella 0.0%
58
Pasteurella 0.0%
59
Lactobacillus 0.0%
My main gut issue is chronic distention/bloat. It hasn't been flat for even one minute for the past several years. It only goes down significantly if I eat very little other wise, I'm always bloated. I don't have any constipation or diahrea. I had two severe episodes of pain where I couldn't stand up straight (literally) several years ago.
I'm very senstive to foods now (in terms of my alleriges/eczema that I've had since I was a kid). Before I could eat a cake made with eggs and be fine (supper allergic to eggs). But now I avoid it strictly and also other things I've become sensitive to: yeast, vinegar, nuts, some fishes.
Anyways I'll just put this out there for anyone who's curious. Any comments are welcomed.
Great! Thanks. You asked, "When you say your skeptical of their ability to determine/predict gut health, are you saying that the results might not be an accurate picture of what the gut looks like?"
DeleteYes, this is exactky what I am saying. One thing, these tests completely miss yeast and fungi, which may actually have more impact than the bacteria. I've looked at probably 100 of these uBiome reports for people, very rarely has a person with big digestive problems had a report that looked considerably different from healthy people.
Maybe uBiome and others will be able to put meaning in these tests when they've collected 1000's of samples, but for now, I think they are meaningless.
My advice for you would be to transition to a very plant-heavy diet and get as much fiber as you can tolerate.
After 2 years of VLC, I became allergic to things I used to eat. Apples, almonds, and cherries would make my eyes swell shut. After 2 years of increased carbs and fiber, I can eat them all I want. Maybe you will find it works for you, too.
That's very interesting what you say about the similarity of the uBiome results you've seen.
DeleteWhen you say "plant-heavy diet" can you be more specific/give me examples? I think I eat a good amount of plant foods, but maybe I can do more. That would help me :)
Go check out Angelo's Plant Paleo plan he is putting together.
Deletehttp://www.humansarenotbroken.com/about/plant-paleo/
It's hard to say what you need or don't need as I do not know what you eat, but most people would benefit by eating lots more overall plant-matter. Things like garlic, onion, leeks, squash, nuts, beans, potatoes, seaweed, etc... should become part of the daily routine.
Thanks Tim for the link, it helped a lot. After thinking about it, I see were I can make improvements to my diet.
DeleteMy diet is for the most part plant-based and whole food, but definitely not perfect. I eat meat infrequently, no dairy or eggs. My usual veggies are broccoli and carrots, kale occasionally. I eat beans really infrequently. I stay away from anything with yeast, so no bread. I still do eat some processed foods (limited amounts, but on reflection, pretty consistently in some form, like, granola bars or those natural brand pop tarts or crackers even (occasional). In hindsight there are times where I could be pretty bad with no control.
I eat oatmeal for breakfast and usually eat lots of sweet potatoes, rice, farro, sometimes quinoa. I think my diet is probably more starch heavy in comparison to my veggie content and my starches could probably be better. I'm probably not consistent as I should be.
After looking at the link I'm going to add more veggies, try to eat some leafy greens daily, eat legumes more. I'm trying to get into the habit of putting the rice in the fridge instead of leaving it on keep warm in the rice cooker. More potatoes. More garlic and onions. I'll have to try leeks, never ate them before.
I've actually been trying to do 100% no processed foods, I always eat at least a bit though so I need a bit of discipline.
I'm going to think more about my diet while trying to implement these things and I've been thinking about keeping a food diary (I first had the thought for allergy purposes).
Sometimes I think it hangs me up that I think my diet is not bad and really good compared to others so it makes me lax in making even better choices. But also I have to remember that my digestive system has some issues and I have to go the extra mile if I want it to heal.
Anyways, thanks for the link. One other thing I'm trying to improve on is sleeping early. I think you commented that that was something you always do and helps your health, am I right?
Dear Tim,
ReplyDeleteI noticed by my stool –diagnostic-tests, which I´m doing all three month, that when I take KS (20-30 g) and fibers (50-60 g) each day, that I have high mark/level on candida albicans or candida species.
Since two years I just eat rice; steamed carrots and parsnips, steamed salmon all boiled in bone chicken soup. The output ;-)) is undigested yellow stool or strong constipation or severe nausea, etc. any inputs/help?
Should I take in my daily meal carbohydrates?
I have CBS and want to treat it with wild oregano oil and berberin, are there any other possibilities?
Any comments would be great!
Thanks